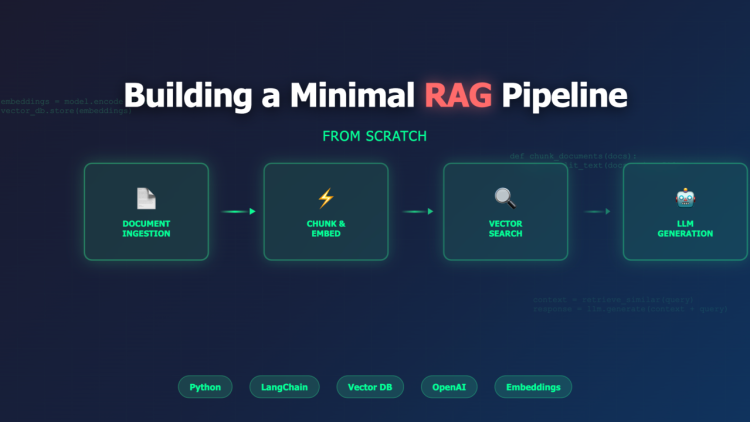
1) Architecture at a glance
[Docs/PDFs/Markdown]
│ ingest + chunk
▼
[Embeddings Model] ──► [Vector Index (FAISS/PgVector)]
▲
│ top-k by query embedding
User Question ──embed──► [Retriever] ──► [Prompt Builder] ──► [LLM]
│
Answer + Sources
2) Minimal stack (why these)
- Chunking: 500–800 tokens with 50–100 overlap → balances recall & context.
- Embeddings: sentence-transformers (local) or OpenAI text-embedding-3-large (managed).
- Index: FAISS for single-machine; switch to pgvector for multi-service.
- LLM: Any function-calling or plain chat model; keep it swappable.
3) The tiniest working example (Python)
Works on a laptop. Replace the model names/keys as you like.
Install
pip install sentence-transformers faiss-cpu pypdf tiktoken openai
3.1 Ingest & chunk
# ingest.py
import os, glob, re
from pypdf import PdfReader
def read_txt(path): return open(path, "r", encoding="utf-8", errors="ignore").read()
def read_pdf(path):
r = PdfReader(path)
return "\n".join(page.extract_text() or "" for page in r.pages)
def load_corpus(folder="docs"):
texts = []
for f in glob.glob(os.path.join(folder, "**/*"), recursive=True):
if os.path.isdir(f): continue
if f.lower().endswith((".md",".txt",".markdown")):
texts.append((f, read_txt(f)))
elif f.lower().endswith(".pdf"):
texts.append((f, read_pdf(f)))
return texts
def chunk(text, size=800, overlap=120):
# rough token-ish splitter on sentences
sents = re.split(r'(?<=[.?!])\s+', text)
chunks, cur = [], []
cur_len = 0
for s in sents:
cur.append(s)
cur_len += len(s.split())
if cur_len >= size:
chunks.append(" ".join(cur))
# overlap
back = " ".join(" ".join(cur).split()[-overlap:])
cur = [back]
cur_len = len(back.split())
if cur:
chunks.append(" ".join(cur))
return chunks
3.2 Build embeddings & FAISS index
#index.py
import faiss, pickle
from sentence_transformers import SentenceTransformer
from ingest import load_corpus, chunk
EMB_NAME = "sentence-transformers/all-MiniLM-L6-v2" # small & fast
def build_index(folder="docs", out_dir="rag_store"):
model = SentenceTransformer(EMB_NAME)
records = [] # [(doc_id, chunk_text, metadata)]
vectors = [] # list of embeddings
for path, text in load_corpus(folder):
for i, ch in enumerate(chunk(text)):
emb = model.encode(ch, normalize_embeddings=True)
vectors.append(emb)
records.append((f"{path}#chunk{i}", ch, {"source": path, "chunk": i}))
dim = len(vectors[0])
index = faiss.IndexFlatIP(dim) # cosine with normalized vectors ~ inner product
import numpy as np
mat = np.vstack(vectors).astype("float32")
index.add(mat)
os.makedirs(out_dir, exist_ok=True)
faiss.write_index(index, f"{out_dir}/index.faiss")
with open(f"{out_dir}/records.pkl", "wb") as f:
pickle.dump(records, f)
if __name__ == "__main__":
build_index()
3.3 Query → retrieve → augment → generate
# query.py
import faiss, pickle, numpy as np, os
from sentence_transformers import SentenceTransformer
from openai import OpenAI
EMB_NAME = "sentence-transformers/all-MiniLM-L6-v2"
STORE = "rag_store"
K = 5
def retrieve(question):
model = SentenceTransformer(EMB_NAME)
q = model.encode(question, normalize_embeddings=True).astype("float32").reshape(1, -1)
index = faiss.read_index(f"{STORE}/index.faiss")
with open(f"{STORE}/records.pkl","rb") as f: records = pickle.load(f)
D, I = index.search(q, K)
hits = [records[i] for i in I[0]]
return hits # [(id, text, meta), ...]
def build_prompt(question, hits):
context = "\n\n".join(
[f"[{i+1}] Source: {h[2]['source']}\n{h[1][:1200]}" for i,h in enumerate(hits)]
)
return f"""You are a precise assistant. Use ONLY the context to answer.
If the answer isn't in the context, say "I don't know" and suggest where to look.
Cite sources like [1], [2] that map to the snippets below.
Question: {question}
Context:
{context}
"""
def answer(question):
hits = retrieve(question)
prompt = build_prompt(question, hits)
# Choose your LLM. Here: OpenAI for example; swap with local server if needed.
client = OpenAI() # requires OPENAI_API_KEY
resp = client.chat.completions.create(
model="gpt-4o-mini",
messages=[{"role":"user","content":prompt}],
temperature=0.2
)
return resp.choices[0].message.content
if __name__ == "__main__":
print(answer("What are the refund steps for premium auctions?"))</code></pre>
What this gives you
- Local embedding + FAISS speed.
- Pluggable LLM.
- Answers that only use retrieved chunks and cite sources.
4) Measuring quality
- Retrieval hit-rate: % of test questions where the gold answer appears in top-k chunks.
- Answer accuracy: exact-match / semantic similarity (e.g., BLEURT/BERTScore) against gold answers.
- Hallucination rate: manual spot checks + “I don’t know” rate (should go up slightly when you tighten guardrails).
- Latency/cost: p50/p95 query time, tokens per answer.
Create a small eval set (20–50 Q/A pairs) from your docs. Run it after any change (new chunking, new embedding model).
5) Guardrails I actually used
- Strict prompt: “Use ONLY the context; else say I don’t know.”
- Chunk citations: attach file & chunk IDs; show them in UI.
- Post-validation: regex/JSON checks for structured answers (IDs, amounts, dates).
- Source diversity: prefer hits from different files to avoid redundancy.
6) Common pitfalls (and fixes)
- Over-chunking (too small): loses semantics → drop to 500–800 tokens with 80 overlap.
- Wrong embeddings: domain jargon suffers → try larger models (e5-large, text-embedding-3-large) for critical domains.
- PDF extraction mess: run a cleanup step (collapse hyphens, fix Unicode), or pre-convert to Markdown with a good parser.
- Query drift: add a quick classifier: “Is this answerable from internal docs?” → if not, escalate or say “I don’t know.”
7) From laptop to production
Option A: Keep FAISS, wrap as a service
- Tiny FastAPI server with /search and /answer.
- Nightly cron to re-ingest.
- Cache by normalized question → reuse the same top-k for 24h.
Option B: Move to Postgres + pgvector
- Pros: transactions, backups, horizontal scaling.
- Schema example:
CREATE TABLE rag_chunks ( id bigserial PRIMARY KEY, source text, chunk_index int, content text, embedding vector(1536) -- match your embedding dim ); -- Vector search SELECT id, source, chunk_index, content FROM rag_chunks ORDER BY embedding <#> $1 -- cosine distance with pgvector LIMIT 5;
Option C: Orchestrate with Temporal (reliable pipelines)
- Workflow: ingest → chunk → embed (batched) → upsert index → smoke test → publish.
- Activity retries, idempotent upserts, metrics on every step.
8) Prompt I ship with (copy/paste)
System:
You answer only from the supplied CONTEXT.
If the answer is missing, reply: "I don't know based on the provided documents."
Always include citations like [1], [2] mapping to sources below.
Be concise and exact.
User:
Question: {{question}}
CONTEXT SNIPPETS:
{{#each snippets}}
[{{@index+1}}] Source: {{this.source}}
{{this.text}}
{{/each}}
9) What I’d add next (nice upgrades)
- Hybrid retrieval: BM25 (keyword) + vectors → better on numbers/code.
- Reranking: small cross-encoder re-rank the top-50 to top-5 (big relevance win).
- Multi-tenant: per-team namespaces, per-doc ACLs.
- Inline quotes: highlight matched spans in each chunk.
- Evals dashboard: store runs in SQLite/Parquet and chart trends.
10) Repo structure (starter)
rag/
docs/ # your source files
rag_store/ # generated index + records
ingest.py
index.py
query.py
evals/
qa.jsonl # [{"q":"..","a":"..","ids":[...]}]
server.py # optional FastAPI
requirements.txt
Final thought
RAG works best when it’s boringly deterministic around a very flexible LLM. Keep the moving parts few, measure retrieval first, and make “I don’t know” an acceptable, logged outcome. Ship tiny, improve weekly.






Discussion about this post